Find False Negatives
In the case of missing variants (not called by the variantCaller plugin), an alignment viewer, such as Integrative Genomics Viewer (IGV) or IGV Light in Ion Reporter™ software, is a valuable tool to verify the presence of the variant in the sample at the position where it is expected.
-
IGV may reveal problems that are not imputable to the variantCaller - for example, problems in mapping or low coverage.
-
Visually inspect the coverage of the region where the variant is expected, paying special attention to the depth of coverage and the quality of the bases covering the position of the variant. Low coverage or low base quality might explain the no-call.
-
The variant could be slightly misplaced (especially for indels).
Optionally, TVC's built-in tools for displaying call details can be used.
If a hotspots file was used:
- Check that the position of the variant is included in the hotspots file.
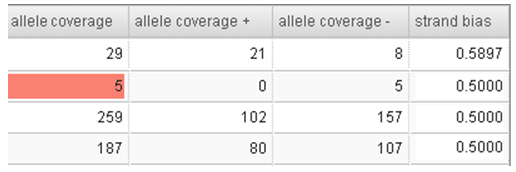
- Check the Variant Calls output table. Values that cause a candidate to be filtered out are shown in colored cells:
- Adjust parameters.
- Run the variant caller again.
If no hotspots file was used:
-
Navigate to the variantCaller results directory on the Torrent Server and open the file small_variants_filtered.vcf. On Linux, the TS variantCaller results directory can be found at /results/analysis/output/Home/{analysis_report_name}/plugins/variantCaller/ for non-barcoded runs or /results/analysis/output/Home/{analysis_report_name}/plugins/variantCaller/{bar code}/ for barcoded runs. In the Torrent Browser, you can access the variantCaller results directory by opening the variantCaller report page for the sample or barcode of interest, removing the final 'variantCaller.html' from the URL, and hit Enter.
-
If the location of the variant is found, look at the FR field (filtered reason).
-
Relate the reason to parameters using the table Filtering Codes variantCaller v4.x.
-
Adjust parameters.
-
Run the variant caller again.
If the location of the SNP is NOT found in the filtered.vcf file create a hotspots file including this location.